Lipid Bilayers
- Details
- Last Updated: Thursday, 11 February 2021 11:23
Many properties of lipid membranes have been characterized over the past decades by all-atom models. Such properties include the area per lipid, order parameters, and density profiles for instance. Currently 100s of nanoseconds are routinely probed for membranes containing a few thousands of lipids.
However, for many membrane related processes more extensive sampling is required, either in the time domain or in terms of an increased system size to study collective events. In those cases the Martini model is a good alternative. Examples include the nucleation of gel domains in either pure [1] or mixed [2] lipid membranes (see figure), transition toward non-lamellar phases [3], partitioning of other compounds such as alcohols [9,19], voltage sensitive dyes [4], phytochemicals [12], the flip-flop of cholesterol [5,6] and the clustering behavior of gangliosides [17]. Furthermore, extensive sampling allowed us to compute the local area compressibility along the membrane normal, an important quantity that had not been considered before [11].
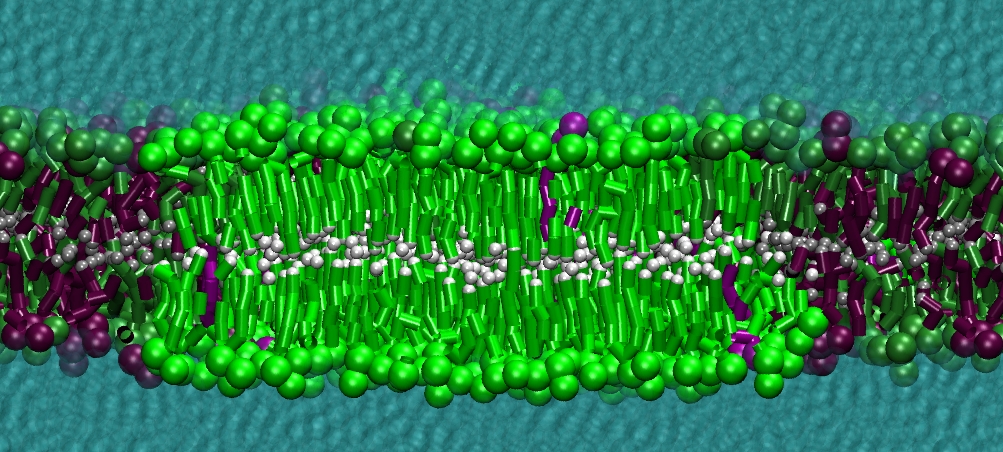
The Martini model can also be efficiently used for systematic studies in which a large number of simulations need to be performed, i.e., in high-throughput applications. For instance, the membrane area and thickness as a function of location of double bonds [7], or the self-diffusion of transmembrane peptides as a function of peptide sequence [8] have been systematically explored with Martini, as well as the in-silico design of membranes with controlled properties using e.g. bolalipids [10]. More recently, the effect of periodic boundary conditions on the lipid self-diffusion has been tested in a series of long timescale and increasing length scale simulations [18] up to patches of more than half a million lipids [22]. In another high-throughput application, Martini was used to study the membrane permeation process of more than 500000 (!) compounds [23].
A number of dedicated tools are available to automatize the initialization of Martini membrane simulations, such as Insane [14] and Charmm GUI Martini-maker [15,16], with hundreds of different lipid types currently available. For even larger scale lipid applications, the Dry martini force field is available [13]. Coupling to all-atom membrane models can be done with our virtual site approach [20,21].
- [1] S.J. Marrink, J. Risselada, A.E. Mark. Simulation of gel phase formation and melting in lipid bilayers using a coarse grained model. Chem. Phys. Lip., 135:223-244, 2005.
- [2] R. Faller, S.J. Marrink. Simulation of domain formation in DLPC-DSPC mixed bilayers. Langmuir, 20:7686-7693, 2004.
- [3] S.J. Marrink, A.E. Mark. Molecular view of hexagonal phase formation in phospholipid membranes. Biophys. J., 87:3894-3900, 2004.
- [4] M.J. Hinner, S.J. Marrink, A.H. de Vries. Location, tilt, and binding: a molecular dynamics study of voltage sensitve dyes in biomembranes. J. Phys. Chem. B, 113:15807-15819, 2009.
- [5] S.J. Marrink, A.H. de Vries, T.A. Harroun, J. Katsaras, S.R. Wassall. Cholesterol shows preference for the interior of polyunsaturated lipid membranes. JACS, 130:10-11, 2008.
- [6] W.F.D. Bennett, J.L. MacCallum, M.J. Hinner, S.J. Marrink, D.P. Tieleman. A molecular view of cholesterol flip-flop and chemical potential in different membrane environments. JACS, 131:12714-12720, 2009.
- [7] N. Kucerka, J. Gallova, D. Uhrikova, P. Balgavy, M. Bulacu, S.J. Marrink, J. Katsaras. Areas of monounsaturated diacylphosphatidylcholines. Biophys. J., 97:1926-1932, 2009.
- [8] S. Ramadurai, A. Holt, L.V. Schäfer, V.V. Krasnikov, D.T.S. Rijkers, S.J. Marrink, J.A. Killian, B. Poolman. Influence of hydrophobic mismatch and amino acid composition on the lateral diffusion of transmembrane peptides. Biophys. J., 99:1447-1454, 2010.
- [9] M. Klacsová, M. Bulacu, N. Kučerka, D. Uhríková, J. Teixeirad, S.J. Marrink, P. Balgavý. The effect of aliphatic alcohols on fluid bilayers in unilamellar DOPC vesicles – a small-angle neutron scattering and molecular dynamics study. BBA Biomembr., 808:2136-2146, 2011. abstract
- [10] M. Bulacu, X. Periole, S.J. Marrink. In-silico design of robust bolalipid membranes, Biomacromol., 13:196–205, 2012. abstract
- [11] F. Campelo, C. Arnarez, S.J. Marrink, M.M. Kozlov. Helfrich model of membrane bending: from Gibbs theory of liquid interfaces to membranes as thick anisotropic elastic layers. Adv. Colloid Interf. Sci., 208:25-33, 2014. abstract
- [12] H.I. Ingólfsson, P. Thakur, K.F. Herold, E.A. Hobart, N.B. Ramsey, X. Periole, D.H. de Jong, M. Zwama, D. Yilmaz, K. Hall, T. Maretzky, H.C. Hemmings, C. Blobel, S.J. Marrink, A. Kocer, J.T. Sack, O.S. Andersen. Phytochemicals perturb membranes and promiscuously alter protein function. ACS Chem. Biol., 9:1788–1798, 2014. abstract
- [13] C. Arnarez, J.J. Uusitalo, M.F. Masman, H.I. Ingólfsson, D.H. de Jong, M.N. Melo, X. Periole, A.H. De Vries, S.J. Marrink. Dry Martini, a coarse-grained force field for lipid membrane simulations with implicit solvent. JCTC, 11:260–275, 2015. abstract
- [14] T.A. Wassenaar, H.I. Ingólfsson, R.A. Böckmann, D.P. Tieleman, S.J. Marrink. Computational lipidomics with insane: a versatile tool for generating custom membranes for molecular simulations. JCTC, 11:2144–2155, 2015. abstract
- [15] Y. Qi, H.I. Ingólfsson, X. Cheng, J. Lee, S.J. Marrink, W. Im. CHARMM-GUI Martini Maker for coarse-grained simulations with the Martini force field. JCTC, 11:4486–4494, 2015. abstract
- [17] R.X. Gu, H.I. Ingólfsson, A.H. de Vries, S.J. Marrink, D.P. Tieleman. Ganglioside-lipid and ganglioside-protein interactions revealed by coarse-grained and atomistic molecular dynamics simulations. JPCB, 121:3262–3275, 2017. open access
- [18] R.M. Venable, H.I. Ingólfsson, M.G. Lerner, B.S. Perrin, Jr., B.A. Camley, S.J. Marrink, F.L.H. Brown, R.W. Pastor. Lipid and peptide diffusion in bilayers: The Saffman-Delbrück model and periodic boundary conditions. JPCB, 121:3443–3457, 2017. abstract
- [20] Y. Liu, W. Pezeshkian, J. Barnoud, A.H. de Vries, S.J. Marrink. Coupling coarse-grained to fine-grained models via Hamiltonian replica exchange. JCTC, 2020. doi.org/10.1021/acs.jctc.0c00429
- [21] Y. Liu, A.H. de Vries, J. Barnoud, W. Pezeshkian, J. Melcr, S.J. Marrink. Dual resolution membrane simulations using virtual sites. JPCB 124:3944, 2020. doi.org/10.1021/acs.jpcb.0c01842
- [22] M. Vögele, J. Köfinger, G. Hummer. Hydrodynamics of Diffusion in Lipid Membrane Simulations. Phys. Rev. Lett. 120:268104, 2018.
- [23] R. Menichetti, K.H. Kanekal, T. Bereau. Drug–Membrane Permeability across Chemical Space. ACS Central Science 5:290-298, 2019.
doi:10.1021/acscentsci.8b00718


























































































































































