Membrane Fusion
- Details
- Last Updated: Thursday, 04 February 2021 16:58
Membrane fusion lies at the heart of important biological processes. In vivo, membrane fusion is tightly regulated by proteins. The basic mechanism, however, is primarily determined by the physics of lipid-lipid interactions.
We have used the Martini model to provide insight into the molecular details of fusion, either between planar lipid membranes [2,4] or between small vesicles [1]. Depending on lipid composition, the rate of fusion might vary by orders of magnitude, and the system can be trapped at any of the intermediate stages of fusion such as the stalk or the hemifusion state.
The effect of peptides on the fusion process can also be studied; for instance, we found that [3,8,9] fusion peptides stabilize a stalk/pore complex, an important intermediate of the fusion pathway. The fusion of vesicles mediated by lung surfactant protein B [5] and the SNARE complex [6] have also been simulated with Martini.
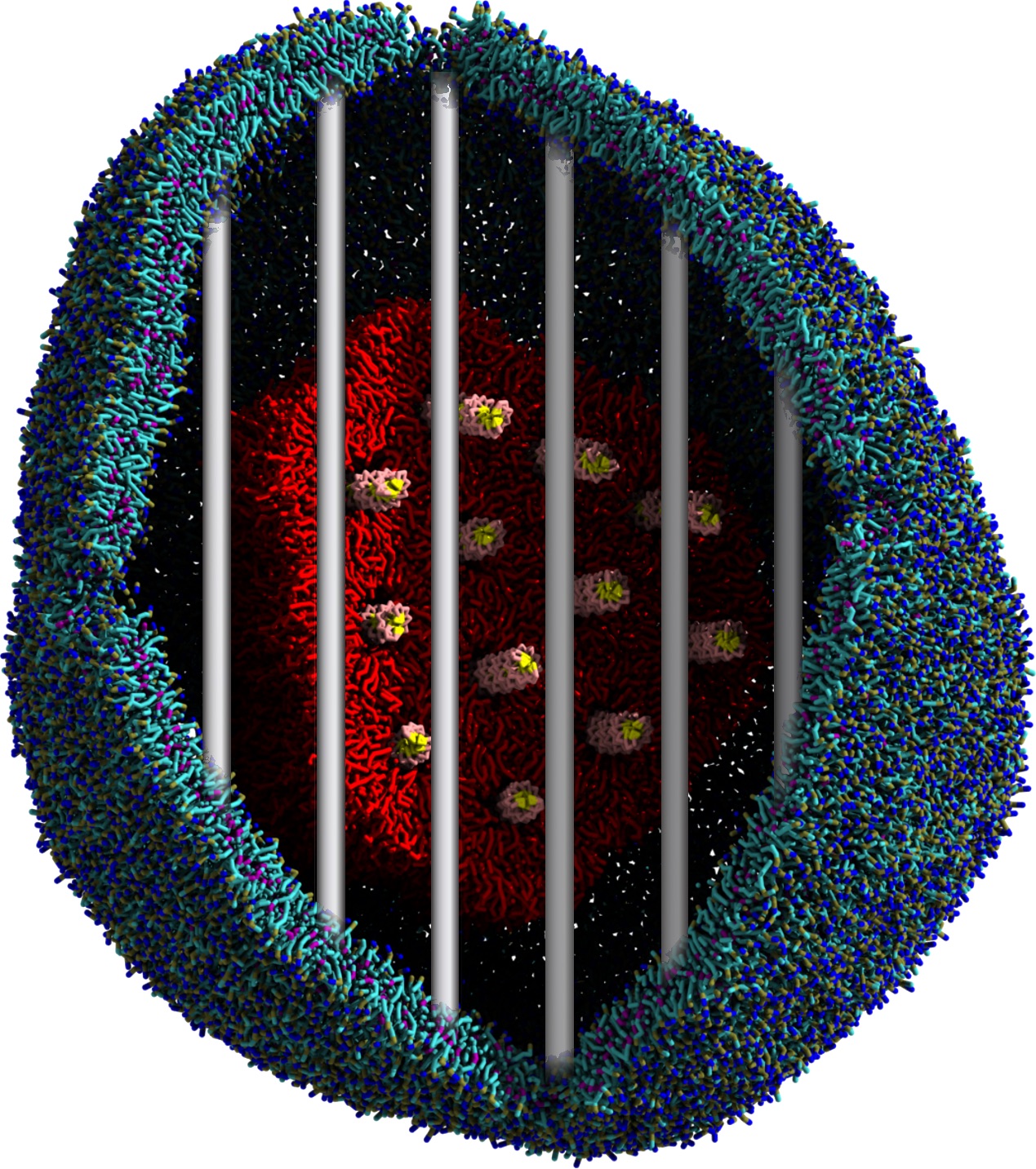
Another study by our lab investigated the mechanism of calicum and PEG mediated fusion [10]. Most recently, we simulated the fusion of lipoplexes with endosomal membrane models [12], see the Figure.
For more simulations on fusion, including many Martini-based works, see [7,11].
[1] S.J. Marrink, A.E. Mark. The mechanism of vesicle fusion as revealed by molecular dynamics simulations. JACS, 125:11144-11145, 2003.
[2] S.J. Marrink, A.E. Mark. Molecular view of hexagonal phase formation in phospholipid membranes. Biophys. J., 87:3894-3900, 2004.
[3] M. Fuhrmans, V. Knecht, S.J. Marrink. A single bicontinuous cubic phase induced by fusion peptides. JACS, 131:9166-9167, 2009.
[4] Y.G. Smirnova, S.J. Marrink, R. Lipowsky, V. Knecht. Solvent-exposed tails as prestalk transition states for membrane fusion at low hydration. JACS, 132:6710-6718, 2010.
[5] S. Baoukina, D.P. Tieleman. Direct simulation of protein-mediated vesicle fusion: lung surfactant protein B. Biophys. J., 99:2134-2142, 2010.
[6] H.J. Risselada, C. Kutzner, H. Grubmüller. Caught in the act: Visualization of SNARE-mediated fusion events in molecular detail. ChemBioChem, 12, 2011.
[7] A.J. Markvoort, S.J. Marrink. Lipid acrobatics in the membrane fusion arena, In Curr. Top. Membr. in Membrane Fusion, Vol. 68, L.V. Chernomordik, M.M. Kozlov Eds. Elsevier, New York, NY, pp. 259-294, 2011. abstract, reprint
[8] M. Fuhrmans, S.J. Marrink. Molecular view of the role of fusion peptides in promoting positive membrane curvature. JACS, 134:1543–1552, 2012. abstract
[9] H.J. Risselada, G. Marelli, M. Fuhrmans, Y.G. Smirnova, H. Grubmüller, S.J. Marrink, M. Muller. Line-tension controlled mechanism for influenza fusion. PLoS ONE 7:e38302, 2012. open access
[10] M. Pannuzzo, D.H. de Jong, A. Raudino, S.J. Marrink. Simulation of polyethylene glycol and calcium-mediated membrane fusion. J. Chem. Phys., 140:124905, 2014. abstract
[11] S.J. Marrink, V. Corradi, P.C.T. Souza, H.I. Ingolfsson, D.P. Tieleman, M.S.P. Sansom. Computational Modeling of Realistic Cell Membranes. Chem. Review, 119:6184–6226, 2019. doi:10.1021/acs.chemrev.8b00460
[12] B.M.H. Bruininks, P.C.T. Souza, H. Ingolfsson, S.J. Marrink. A molecular view on the escape of lipoplexed DNA from the endosome. eLife, 9:e52012, 2020. doi.org/10.7554/eLife.52012


























































































































































