Realistic Cell Membranes
- Details
- Last Updated: Thursday, 18 February 2021 16:53
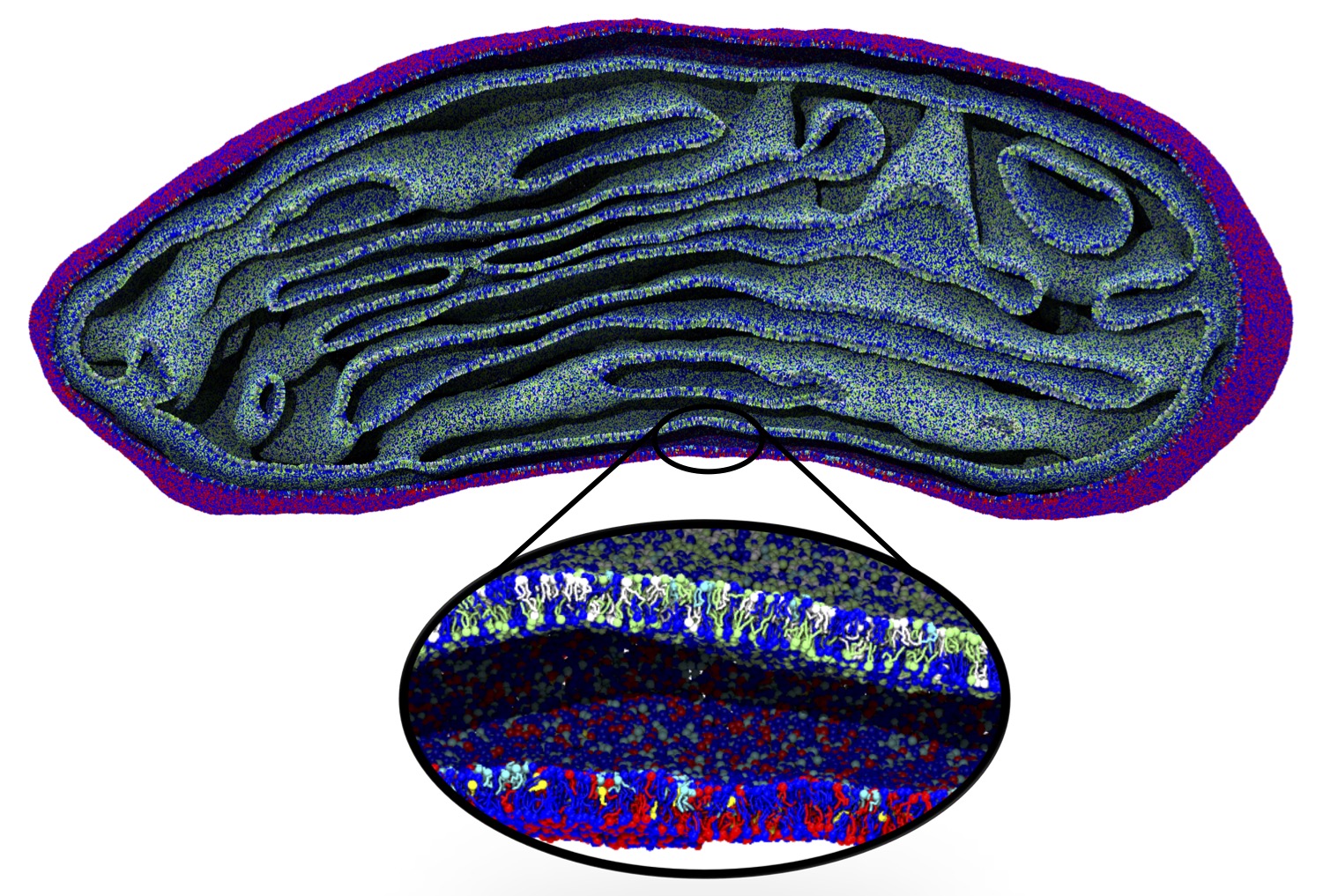 Biomembranes are essential cellular components. Together with membrane-adhered structures, such as the cytoskeleton, cell membranes constitute incredibly heterogeneous and crowded environments, containing hundreds of different lipid types and being densely packed with a large variety of membrane proteins. They provide identity not only to the cell as a whole, through the enveloping plasma membrane, but also to many internal organelles.
Biomembranes are essential cellular components. Together with membrane-adhered structures, such as the cytoskeleton, cell membranes constitute incredibly heterogeneous and crowded environments, containing hundreds of different lipid types and being densely packed with a large variety of membrane proteins. They provide identity not only to the cell as a whole, through the enveloping plasma membrane, but also to many internal organelles.
The Martini model is highly suited to capture the complexity of such cell membranes, as recently reviewed in [1]. Available tools such as Insane [2] and Charmm-GUI [3,4] facilitate setting up cell membranes with arbitrary complex compositions. Current highlights include the >60 lipid mixture representing mammalian plasma membranes [5-7], mixtures of galactolipids modeling the thylakoid membranes [8], the membranes of an enitre mitochondrion with realistic composition as well as shape [9, see Figure], and multi-component bacterial membranes [10].
The ability to simulate membranes with realistic lipid compositions is now enabling researchers to study the interaction of a variety of proteins and other compounds with such membranes, e.g. [11-14].
[1] S.J. Marrink, V. Corradi, P.C.T. Souza, H.I. Ingolfsson, D.P. Tieleman, M.S.P. Sansom. Computational Modeling of Realistic Cell Membranes. Chem. Review, 119:6184–6226, 2019. doi:10.1021/acs.chemrev.8b00460
[2] T.A. Wassenaar, H.I. Ingólfsson, R.A. Böckmann, D.P. Tieleman, S.J. Marrink. Computational lipidomics with insane: a versatile tool for generating custom membranes for molecular simulations. JCTC, 11:2144–2155, 2015. abstract
[3] Y. Qi, H.I. Ingólfsson, X. Cheng, J. Lee, S.J. Marrink, W. Im. CHARMM-GUI Martini Maker for coarse-grained simulations with the Martini force field. JCTC, 11:4486–4494, 2015. abstract
[5] H.I. Ingólfsson, M.N. Melo, F.J. van Eerden, C. Arnarez, C.A. López, T.A. Wassenaar, X. Periole, A.H. De Vries, D.P. Tieleman, S.J. Marrink. Lipid organization of the plasma membrane. JACS, 136:14554-14559, 2014. open access
[6] H.I. Ingólfsson, T.S. Carpenter, H. Bhatia, P.T. Bremer, S.J. Marrink, F.C. Lightstone. Computational Lipidomics of the Neuronal Plasma Membrane. Biophys. J. 113:2271–2280, 2017. open access
[7] S. Thallmair, H.I. Ingólfsson, S.J. Marrink. Cholesterol Flip-Flop Impacts Domain Registration in Plasma Membrane Models. J. Phys. Chem. Lett. 9:5527–5533, 2018. doi:10.1021/acs.jpclett.8b01877
[8] F.J. van Eerden, D.H. de Jong, A.H de Vries, T.A. Wassenaar, S.J. Marrink. Characterization of thylakoid lipid membranes from cyanobacteria and higher plants by molecular dynamics simulations. BBA Biomembranes, 1848:1319–1330, 2015. abstract
[9] W. Pezeshkian, M. Konig, T.A. Wassenaar, S.J. Marrink. Backmapping triangulated surfaces to coarse-grained membrane models. Nature Commun. 11:2296, 2020. doi.org/10.1038/s41467-020-16094-y
[10] P.C. Hsu, F. Samsudin, J. Shearer, S. Khalid. It Is Complicated: Curvature, Diffusion, and Lipid Sorting within the Two Membranes of Escherichia coli. JPC-Lett. 8 (22), 5513-5518, 2017.
[11] V. Corradi, E. Mendez-Villuendas, H.I. Ingólfsson, R.X. Gu, I. Siuda, M.N. Melo, A. Moussatova, L.J. DeGagné, B.I. Sejdiu, G. Singh, T.A. Wassenaar, K. Delgado Magnero, S.J. Marrink, D.P. Tieleman. Lipid–Protein Interactions Are Unique Fingerprints for Membrane Proteins. ACS Central Science 4:709–717, 2018. doi:10.1021/acscentsci.8b00143
[12] S. Thallmair, P.A. Vainikka, S.J. Marrink. Lipid Fingerprints and Cofactor Dynamics of Light-Harvesting Complex II in Different Membranes. Biophys. J., 116:1446-1455, 2019. doi:10.1016/j.bpj.2019.03.009
[13] J. Shearer, D. Jefferies, S .Khalid. Outer membrane proteins OmpA, FhuA, OmpF, EstA, BtuB, and OmpX have unique lipopolysaccharide fingerprints. J. Chemical Theory and Computation 15 (4), 2608-2619, 2019.
[14] A. Buyan, C.D. Cox, J. Barnoud, J. Li, H.S.M. Chan, B. Martinac, S.J. Marrink, B. Corry. Piezo1 forms specific, functionally important interactions with phosphoinositides and cholesterol. Biophys. J. 119:1683-1697, 2020. doi.10.1016/j.bpj.2020.07.043


























































































































































