ElNeDyn
- Details
- Last Updated: Thursday, 22 August 2013 10:01
ElNeDyn stands for Elastic Network in Dynamics. In this approach an elastic network (EN; a set of springs or harmonic bonds between interacting sites) is used as a structural scaffold to describe and control the overall shape of a molecule. The EN is then combined with a coarse-grained molecular force field to describe the intra- and inter-molecular interactions.
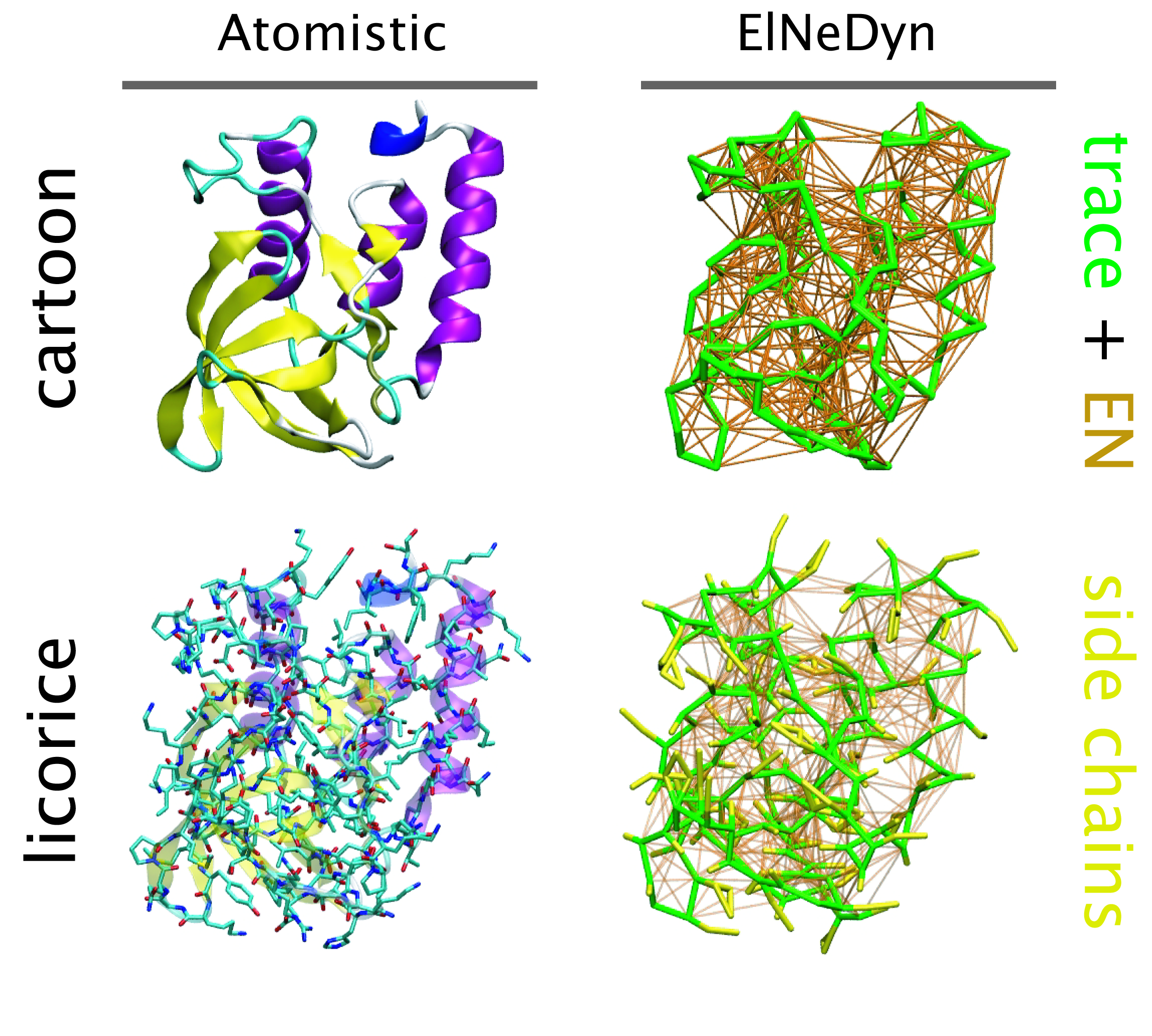 The current version of ElNeDyn focusses on modeling proteins and has been developed in conjunction with the Martini CG force field (2.0, 2.1) but can in principle be mixed with any CG model for any type of molecule. The Martini FF presents the great advantage to include a large body of biomolecules and solvents making simulations of biomolecular systems actually possible.
The current version of ElNeDyn focusses on modeling proteins and has been developed in conjunction with the Martini CG force field (2.0, 2.1) but can in principle be mixed with any CG model for any type of molecule. The Martini FF presents the great advantage to include a large body of biomolecules and solvents making simulations of biomolecular systems actually possible.
The rationale for the use of EN is to control the conformation of the protein while keeping its internal dynamics. This has been possible by parametrizing the EN against atomistic model simulations. By tuning the force constant of the springs and the extend of the network, backbone deformations, fluctuations and large amplitude domain motions observed in atomistic simulations of proteins have been matched. For more detail refer to X. Periole et al. 2009 JCTC 5:2531-43.
One notable detail is that the description of the bonded terms defined in original Martini for proteins has been slightly modified to accommodate for the placement of the backbone bead on the Calpha instead on the center-of-mass of the backbone atoms. The non-bonded terms were used as in the original force field.
In the following you'll find a tar.gz file containing the scripts and programs (listed below) that will allow you to build the topology of protein from a pdb file. The file HowTo-ElNeDyn.pdf describes the steps to follow and a tutorial ia available here.
Download: ElNeDyn-2.1.tar.gz
Files and short description:
- AA.dat : contains the definition of the amino acids.
- cg-2.1.dat : contains the definition of the bonded terms.
- cg-2.1.dat.comments : description of the content of cg-2.1.dat.
- pdb2CGpdb-2.1.f : fortran code reading a pdb file of a protein and giving a CG version of it.
- topol-CG-2.1.f : fortran code reading the CG coordinates and building the ElNeDyn topology.
- HowTo-ElNeDyn.pdf : a step-by-step description of the procedure to get from the pdb file to an MD run.
- a set of mdp files to minimize, relax and simulate your ElNeDyn model.
For more information contact Xavier Periole (This email address is being protected from spambots. You need JavaScript enabled to view it.).
The development of ElNeDyn is done in close collaboration with Prof. M. A. Ceruso at the CCNY-CUNY in New York City, USA.


























































































































































