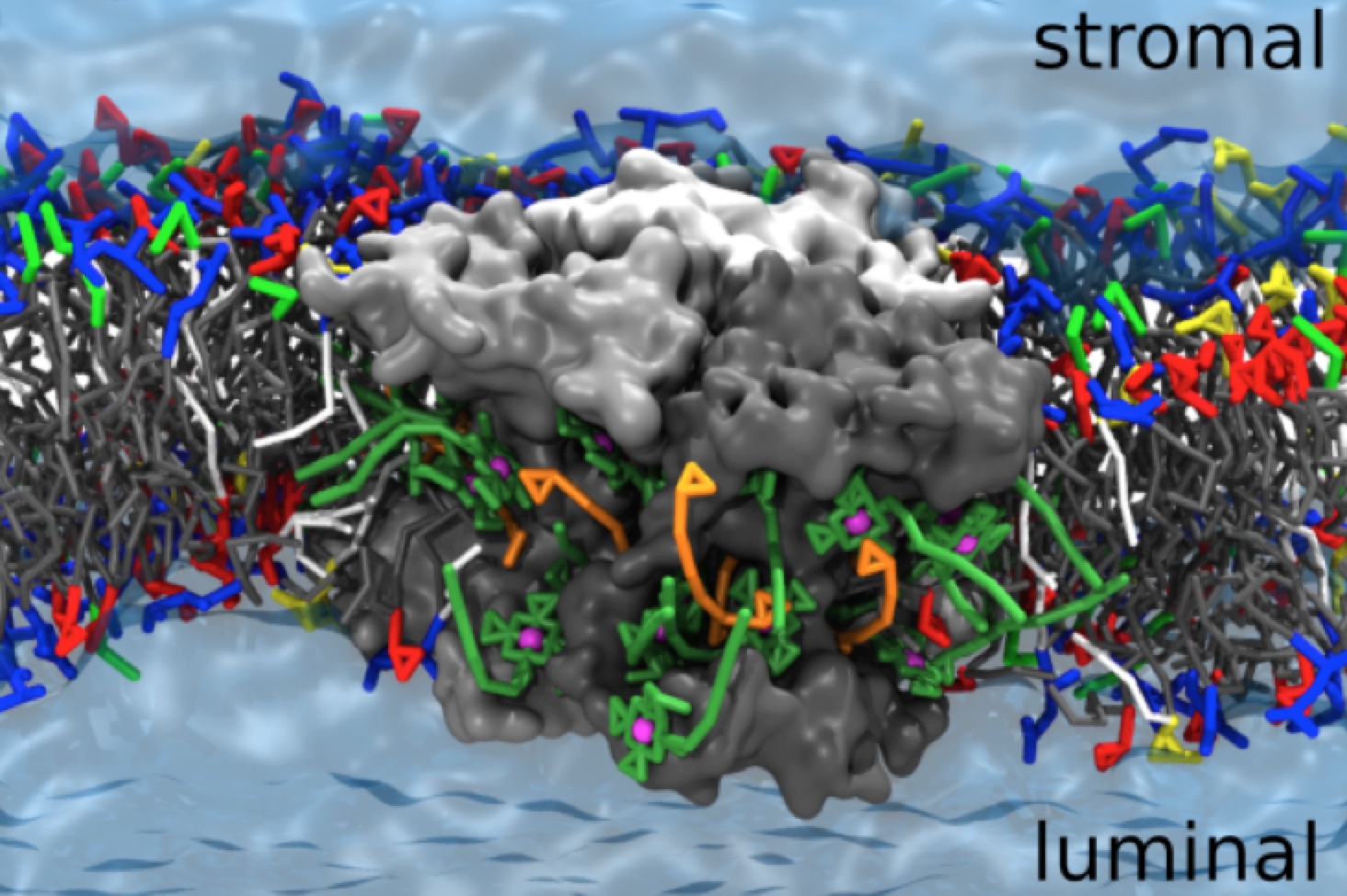
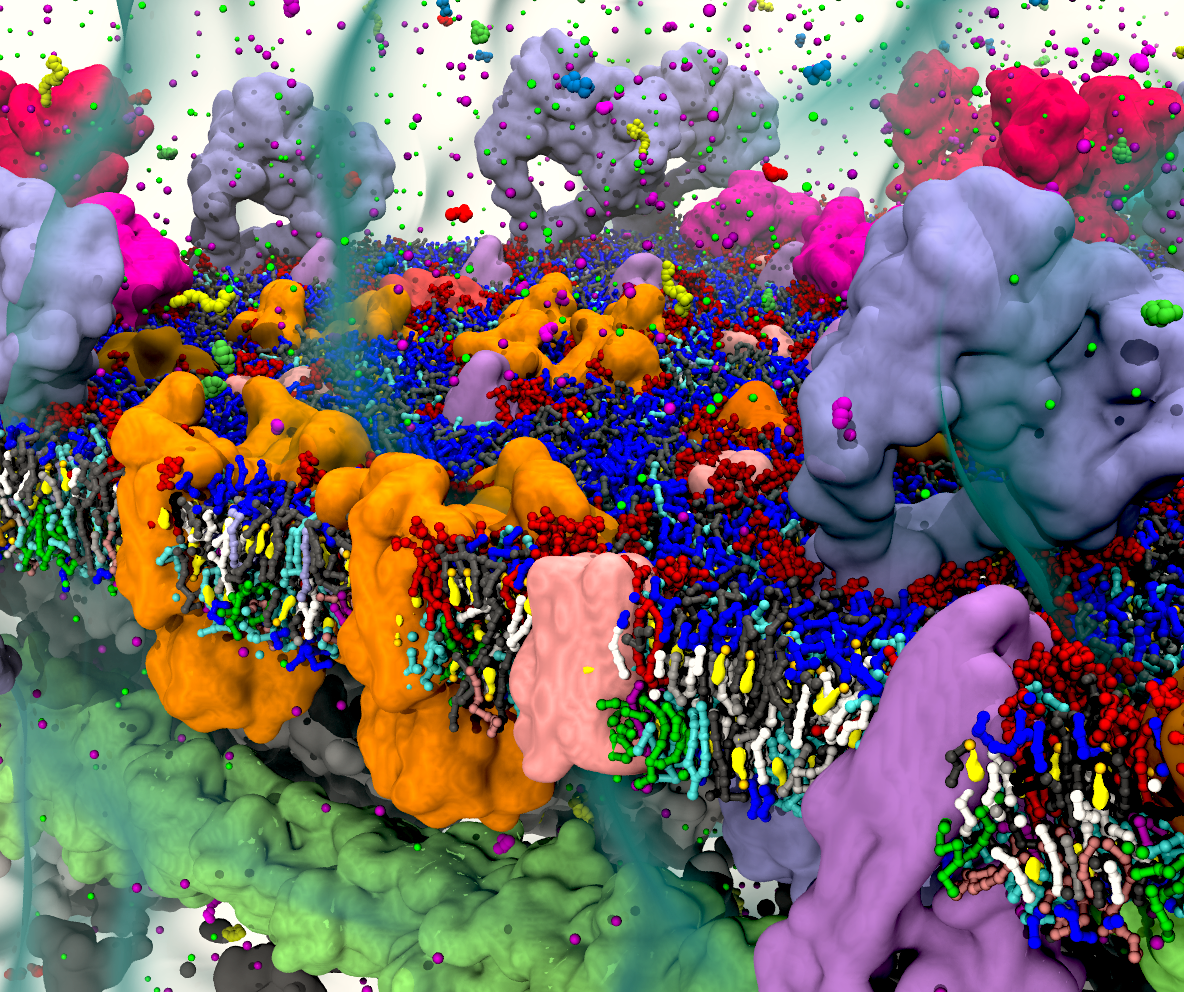
Swinger
- Details
- Last Updated: Saturday, 20 April 2019 19:56
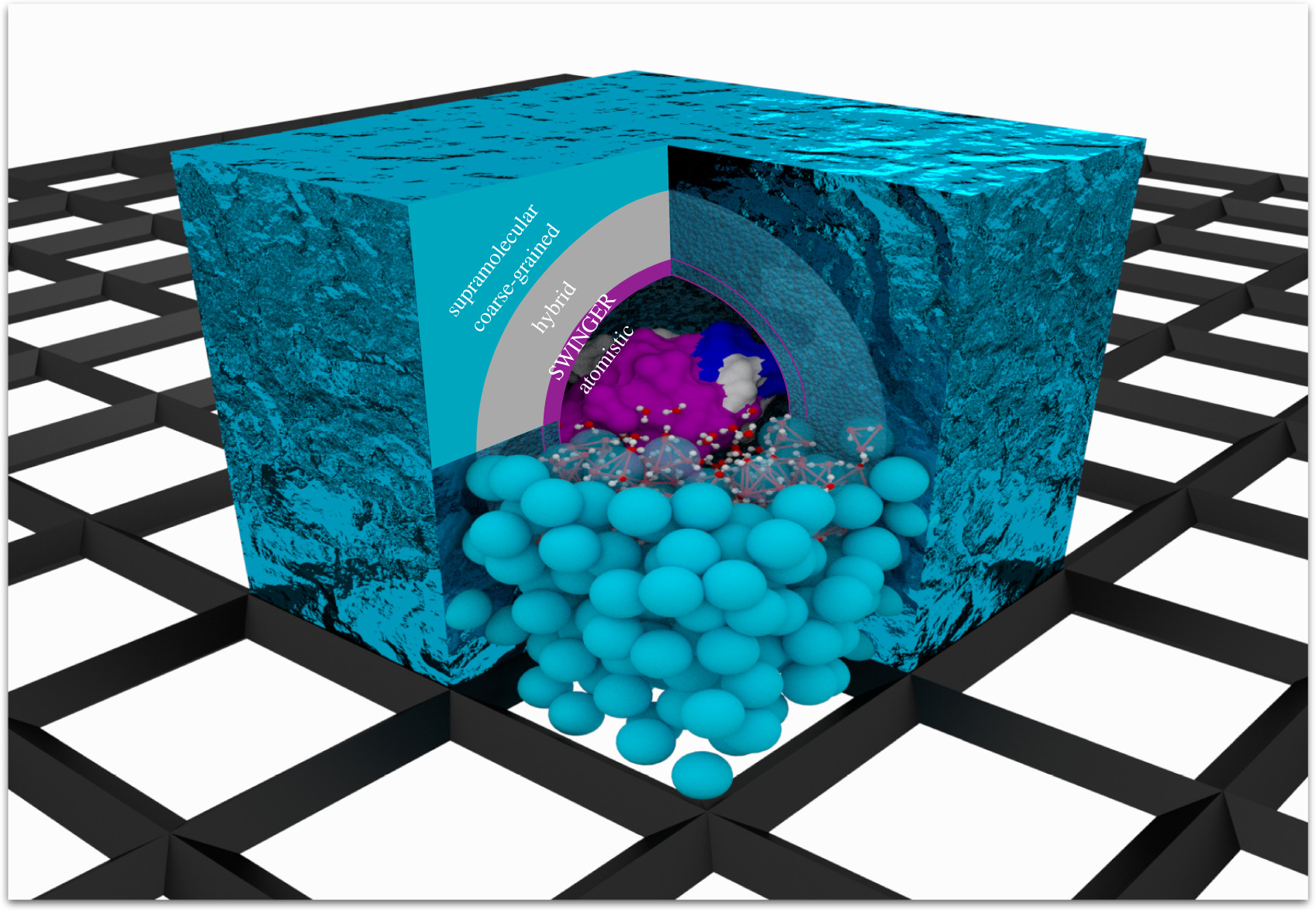
Happy Easter with lots of swinging activities .... For inspiration, check out how to convert your CG water beads to real ones on-the-fly !
J. Zavadlav, S.J. Marrink M. Praprotnik, SWINGER: a clustering algorithm for concurrent coupling of atomistic and supramolecular liquids. Interface Focus 9:20180075, 2019. doi.org/10.1098/rsfs.2018.0075