Martini DNA
- Details
- Last Updated: Thursday, 25 January 2018 13:28
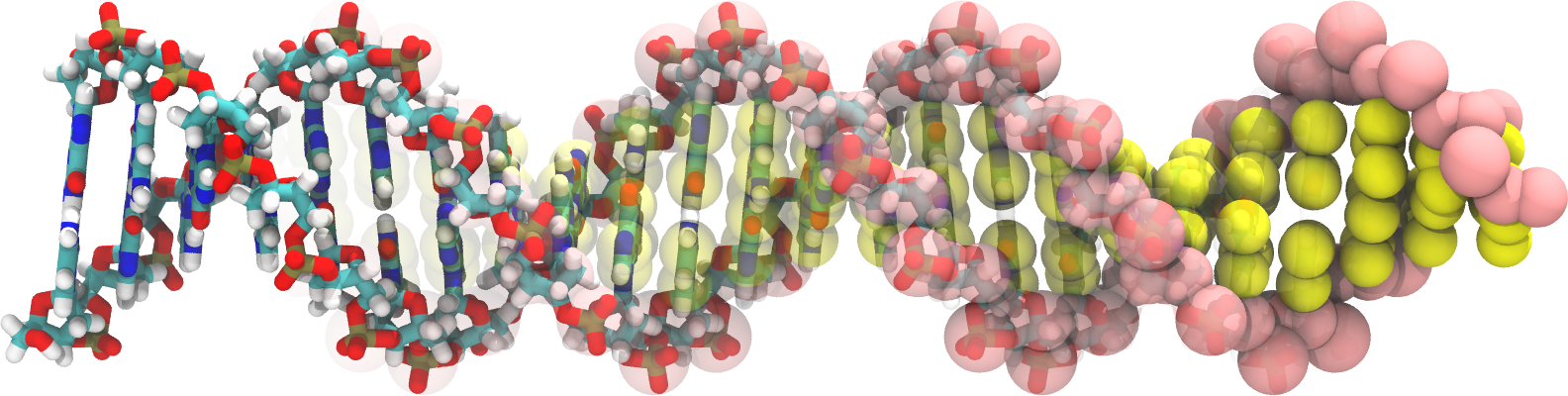
Martini DNA parameters are available as a package with a martinize-dna script to generate Martini topology files for DNA.
An example of an all atomistic 24 bp dsDNA in pdb format, together with its course grained mapping and bonded parameters, using the stiff elastic network settings (20180125).
24bp_AA2CG_stiff.tar.gz
- martini_v2.1P-dna.itp: Fixed missing T-bead - S-bead interactions.
- dna_backmapping_files: Fixed a bug in CHARMM36 thymine files.
- martinize-dna.py: Changed the default cut-off for separating chains to 4 Angstroms.
martini-dna-150722.tgz
For details see:
J.J. Uusitalo, H.I. Ingólfsson, P. Akhshi, D.P. Tieleman, S.J. Marrink
Martini coarse-grained force field: extension to DNA, JCTC (doi: 10.1021/acs.jctc.5b00286)


























































































































































