Martini workshop 2021
- Details
-
Last Updated: Monday, 26 July 2021 22:12
Martini 3.0 online workshop - 1-3 September 2021.
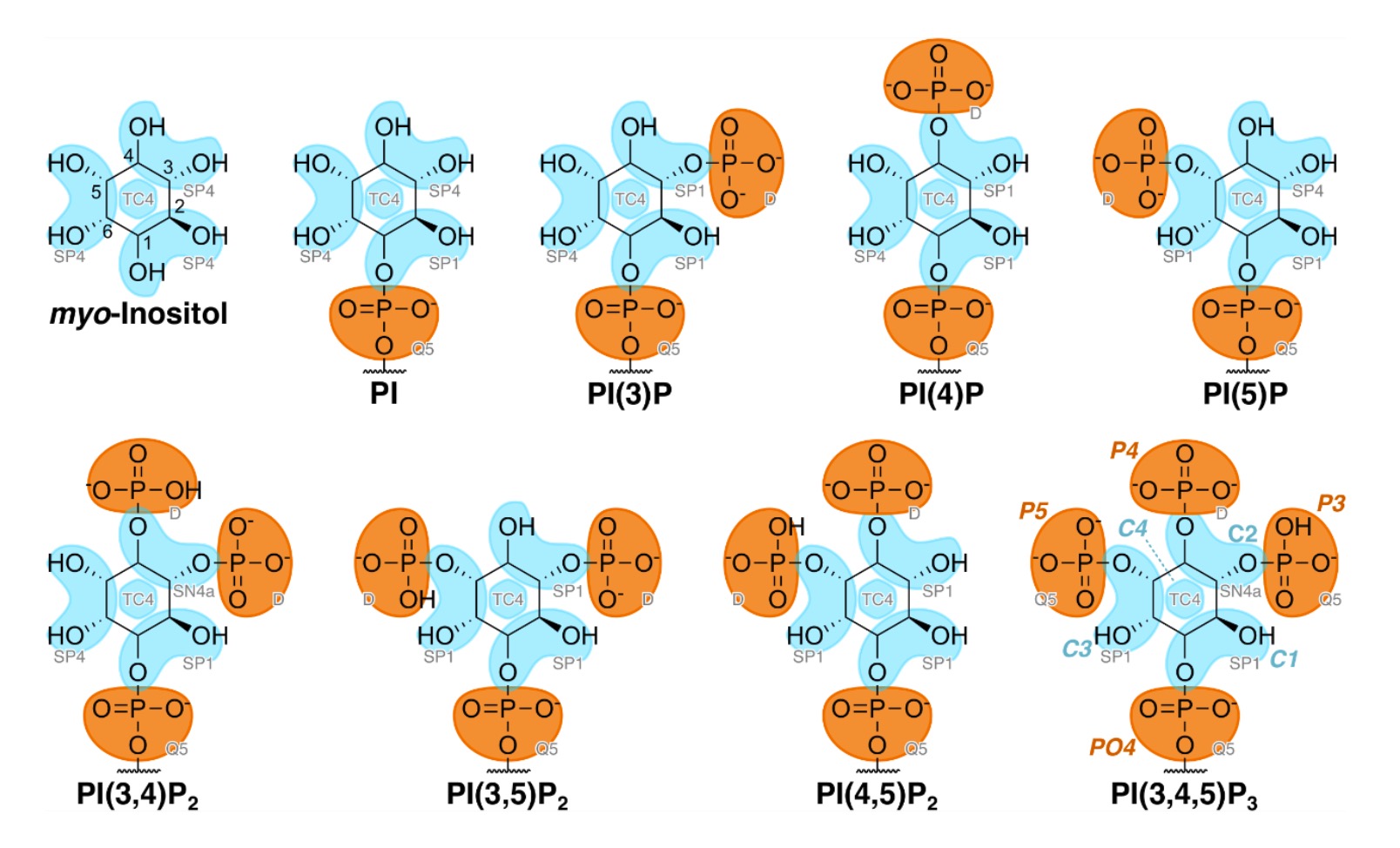
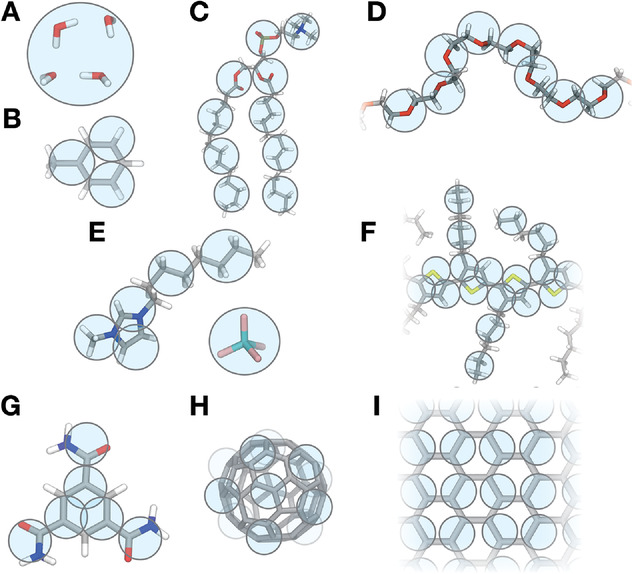
We are happy to announce the next Martini workshop, which will take place online as traveling options are still limited. The three day workshop will consist of a number of pre-recorded lectures and associated Q&A session by some of the key Martini developers, as well as dedicated tutorial sessions where students can practice both basic and advanced Martini simulations under the guidance of expertised assistants. Topics we will cover include, of course, the ins and outs of the latest Martini version, Martini 3.0, and the growing list of associated tools and methods such as martinize2, insane, backwards, TS2CG, polyply, titratable Martini and the Martini database.
Registration for the event is now closed!
Looking forward to meeting you online !
Paulo, Manuel, & Siewert-Jan
(for further questions, please contact Paulo at This email address is being protected from spambots. You need JavaScript enabled to view it.)