Martini MOF
- Details
- Last Updated: Wednesday, 24 May 2023 06:03
Martini keeps expanding its application horizon - see the birth of Martini models for a zeolite MOF: https://doi.org/10.1063/5.0145924
 General Purpose Coarse-Grained Force Field
General Purpose Coarse-Grained Force Field 

























































































































































Martini keeps expanding its application horizon - see the birth of Martini models for a zeolite MOF: https://doi.org/10.1063/5.0145924
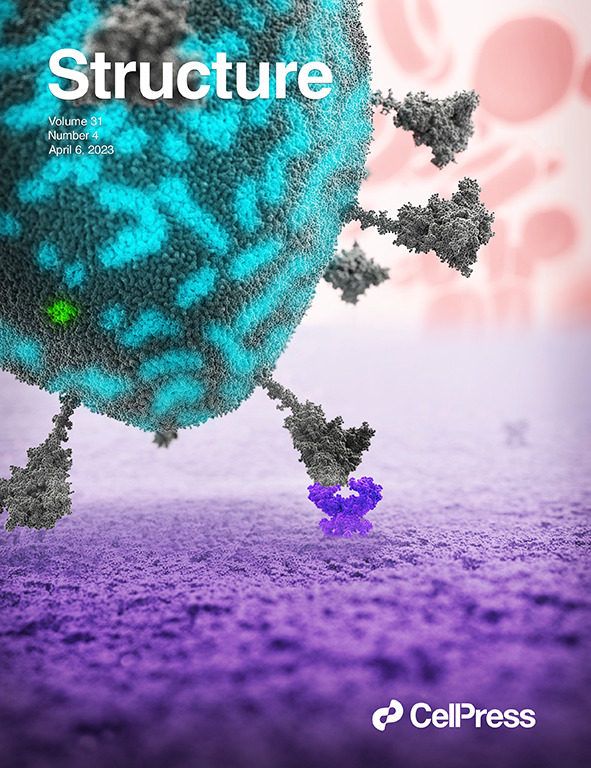
Finally our integrative model of SARS-Cov-2 is published, representing the most realistic model of this viral envelope to date, with interesting pattern formation of the M-dimers.
For details, see Pezeshkian et al., Structure (2023).
Dear users,
Over the past week we have experienced technical difficulties, due to which new users did not receive their registration emails. In case you tried to create an account on this website within the past 10 days, please resubmit the registration application. Apologies for the inconvenience.
Now available: optimized Martini 2 cholesterol model which remedies the artificial temperature gradients using loose LINCS settings. See https://doi.org/10.1021/acs.j
To facilitate the use of Martini in novel application areas, we have published two chapters with practical tips and guidelines on how to (i) setup and run protein-ligand binding simulations, and (ii) use titratable Martini. Enjoy !
The paper presenting MAD - the Martini Database Server - is now online in JCIM: https://pubs.acs.org/doi/abs/10.1021/acs.jcim.2c01375
MAD is designed for the sharing of structures and topologies of molecules parametrized with Martini. MAD also provides a GUI to convert atomistic structures into CG structures and prepare complex systems. Please check it out: https://mad.ibcp.fr