Self-assembling nanotubes
- Details
- Last Updated: Friday, 06 October 2023 15:42
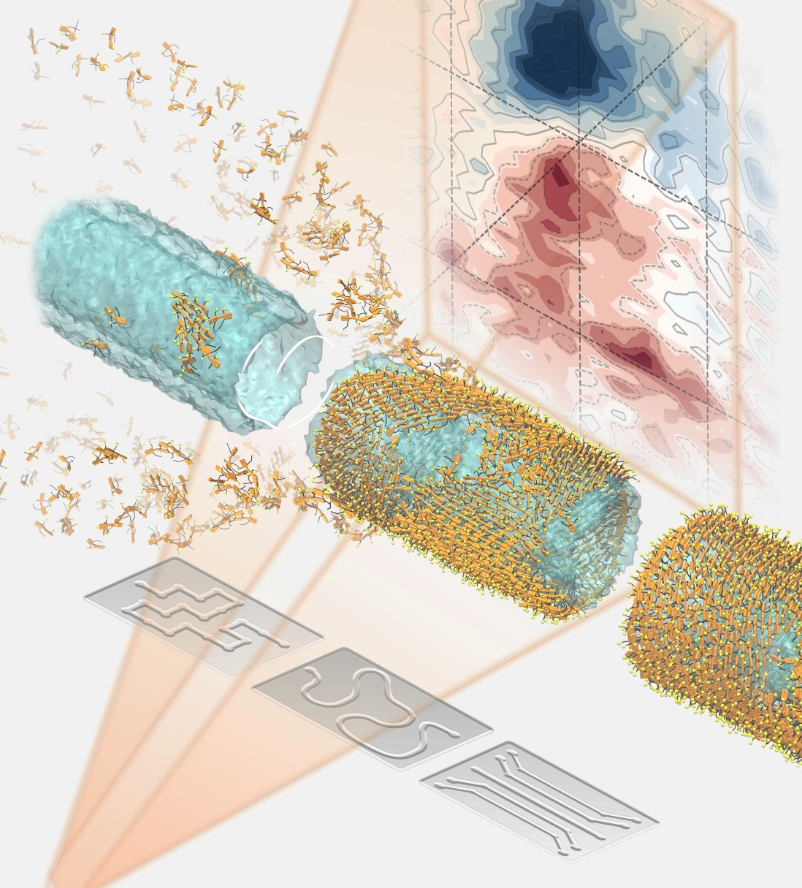
"Watching Molecular Nanotubes Self-Assemble in Real Time", JACS online pubs.acs.org/doi/10.1021/ja Visualization by Marìck Manrho
 General Purpose Coarse-Grained Force Field
General Purpose Coarse-Grained Force Field 


























































































































































"Watching Molecular Nanotubes Self-Assemble in Real Time", JACS online pubs.acs.org/doi/10.1021/ja Visualization by Marìck Manrho
New models for lipopolysaccharides are now available for Martini 3 ! See the paper from Vaiwala and Ayappa, https://doi.org/10.1021/acs.
In MD simulations using the GROMACS package with default Martini parameters, the group of Hummer found large membranes to deform under the action of a semi-isotropically coupled barostat. As the primary cause, they identified overly short outer cutoffs and infrequent neighbor list updates that result in missed long-range attractive Lennard-Jones interactions. Small but systematic imbalances in the apparent pressure tensor then induce unphysical asymmetric box deformations that crumple the membrane. Guidelines are presented to avoid these artefacts, including estimates for appropriate values for the outer cutoff r_l and the number of time steps nstlist between neighbor list updates.
Must-read for those who simulate membrane systems. For details, see the paper from Kim et al. in chemRxiv
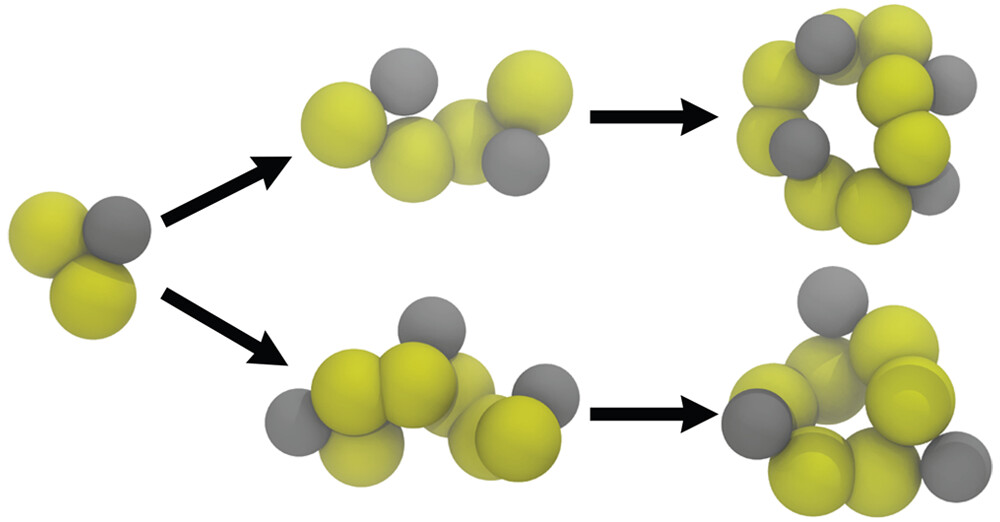
Chemical reactions with Martini ?? Indeed !!
See the paper by Selim Sami, explaining a pragmatic way to capture chemical reactivity in Martini with the clever use of virtual sites: https://doi.org/10.1021/acs.jctc.2c01186
All the required input files together with tutorials on its use are provided as SI.
González, A., Covarrubias-Pinto, A., Bhaskara, R. M., Glogger, M., Kuncha, S. K., Xavier, A., Seemann, E., Misra, M., Hoffmann, M. E., Bräuning, B., Balakrishnan, A., Qualmann, B., Dötsch, V., Schulman, B. A., Kessels, M. M., Hübner, C. A., Heilemann, M., Hummer, G., & Dikić, I. (2023). Ubiquitination regulates ER-phagy and remodelling of endoplasmic reticulum. Nature, 1-8. https://doi.org/10.1038/s41586-023-06089-2
Foronda, H., Fu, Y., Covarrubias-Pinto, A., Bocker, H. T., González, A., Seemann, E., Franzka, P., Bock, A., Bhaskara, R. M., Liebmann, L., Hoffmann, M. E., Katona, I., Koch, N., Weis, J., Kurth, I., Gleeson, J. G., Reggiori, F., Hummer, G., Kessels, M. M., Qualmann, B., . . . Hübner, C. A. (2023). Heteromeric clusters of ubiquitinated ER-shaping proteins drive ER-phagy. Nature, 1-9.https://doi.org/10.1038/s41586-023-06090-9
The group of Tieleman has completed an implementation of Martini 2 and Martini 3 in the OpenMM molecular dynamics software package. OpenMM is a flexible molecular dynamics package widely used for methods development and is competitive in speed on GPUs with other commonly used packages. OpenMM has facilities to easily implement new force field terms, external forces and fields, and other non-standard features, which were used to implement all force field terms of Martini 2 and Martini 3. This allows OpenMM simulations, starting with GROMACS topology files that are processed by custom scripts, with all the added flexibility of OpenMM. For details, see the paper of MacCallum et al., Biophys J., 2023.